Diving into the world of statistics and eager to learn how to run moderation analysis in R with a single moderator? You’ve landed in the right place! Moderation analysis is an important statistical method that allows us to examine and elucidate the complex interplay between variables in our datasets.
However, if you are dealing with multiple moderators, check our comprehensive guide on How To Run Multiple Moderation Analyses in R like a pro.
This comprehensive guide will equip you with the skills to successfully run a moderation analysis in R, a powerful language widely used for statistical analysis. You’ll get hands-on experience with real data and learn to interpret the results within an academic research context.
Lesson Outcomes
By the end of this article, you will have:
- Understand the basic concept of moderation analysis and its role in statistical research.
- Learn how to prepare your data for moderation analysis.
- Familiarize yourself with the necessary R packages and their functions for performing a moderation analysis.
- Gain the ability to conduct a moderation analysis in R, using a linear regression model with an interaction term.
- Learn how to interpret the output of a moderation analysis, including the coefficients of the interaction term.
- Acquire the knowledge to test and validate the assumptions of the linear regression model used in the moderation analysis, including linearity, independence of residuals, homoscedasticity, normality of residuals, and absence of multicollinearity.
- Learn to identify and handle potential outliers and influential observations in your data.
- Apply visual diagnostic tools to assess the fit and assumptions of your model.
- Understand how to report the results of a moderation analysis for academic purposes.
So, if you’re intrigued by the mysteries your data may be hiding and are ready to unravel them, let’s delve into moderation analysis with R together!
What is Moderation Analysis?
Moderation analysis is an analytical method frequently utilized in statistical research to examine the conditional effects of an independent variable on a dependent variable. In simpler terms, it assesses how a third variable, known as a moderator, alters the relationship between the cause (independent variable) and the outcome (dependent variable).
In the language of statistics, this relationship can be described using a moderated multiple regression equation:
Y = \beta_{0} + \beta_{1}X + \beta_{2}Z + \beta_{3}XZ + \epsilon
Where:
- Y represents the dependent variable
- X is the independent variable
- Z symbolizes the moderator variable
- β0, β1, β2, and β3 denote the coefficients representing the intercept, the effect of the independent variable, the effect of the moderator, and the interaction effect between the independent variable and the moderator respectively
- ε stands for the error term
In moderation analysis, the key component is the interaction term (β3*XZ). If the coefficient β3 is statistically significant (p-value <0.05), it indicates the presence of a moderation effect.
Moderation Analysis Example
Let’s assume we are trying to understand if the effect of drinking coffee (X) on productivity (Y) changes based on another factor, like the amount of sleep someone got the night before (Z). We suspect drinking coffee might make people more productive, but perhaps this effect is stronger for those who earn less sleep.
The interaction term (β3*XZ) is essentially saying, “Let’s multiply the number of coffee cups you drink (X) with the amount of sleep you had (Z)” – it’s a new variable that represents the combined effect of coffee and sleep on productivity.
β3 is just the number that shows how much productivity changes when you drink coffee, considering the effect of the amount of sleep. If β3 is statistically significant, it means the amount of sleep does change the relationship between coffee and productivity. Simply put, it’s like saying, “Yes, the effect of coffee on productivity does depend on how much sleep you got!”
If β3 is not statistically significant (p-value <0.05), it would suggest that the amount of sleep doesn’t really matter, and the effect of coffee on productivity is the same regardless of how much sleep you got.
So why do we need moderation analysis? The value of moderation analysis lies in its ability to offer nuanced insights into complex relationships. Instead of investigating direct relationships between variables, moderation analysis allows us to understand ‘under what conditions‘ or ‘for whom‘ these relationships hold. It helps us answer questions more aligned with the complexities of the real-world scenario.
Assumptions of Moderation Analysis
Several assumptions need to be met when conducting moderation analysis. These assumptions are similar to those for multiple linear regression, given that moderation analysis typically involves multiple regression where an interaction term is included. Here are the main assumptions:
- Linearity: The relationship between each predictor (independent variable and moderator) and the outcome (dependent variable) is linear. This assumption can be checked visually using scatter plots we will generate and explain in-depth in this article.
- Independence of observations: The observations are assumed to be independent of each other. This is more of a study design issue than something that can be tested for. If your data is time series or clustered, this assumption is likely violated.
- Homoscedasticity: This refers to the assumption that the variance of errors is constant across all levels of the independent variables. In other words, the spread of residuals should be approximately the same across all predicted values. This can be checked by looking at a plot of residuals versus predicted values.
- Normality of residuals: The residuals (errors) are assumed to follow a normal distribution. This can be checked using a Q-Q plot.
- No multicollinearity: The independent variable and the moderator should not be highly correlated. High correlation (multicollinearity) can inflate the variance of the regression coefficients and make the estimates very sensitive to minor changes in the model. Variance inflation factor (VIF) is often used to check for multicollinearity.
- No influential cases: The analysis should not be overly influenced by any single observation. Cook’s distance can be used to check for influential cases that might unduly influence the estimation of the regression coefficients.
Formulate The Moderation Analysis Model
Let’s assume that we are searching for the answer to the following research question:
“How does the relationship between coffee consumption (measured by the number of cups consumed) and employee productivity vary by individual tolerance to caffeine?“
Therefore we we can formulate the following hypothesis:
“The level of an individual’s caffeine tolerance moderates the relationship between coffee consumption and productivity.“
What we are essentially trying to answer with this hypothesis is whether the impact of coffee consumption on productivity is the same for all individuals, or if it changes based on their caffeine tolerance. In other words, we are investigating if the productivity benefits (or drawbacks) of coffee are the same for everyone, or if they differ based on how tolerant a person is to caffeine.
Therefore, our study consists of the following variables: coffee consumption (independent variable), productivity (dependent variable), and caffeine tolerance (the moderator).
The following diagram explains the type of variables in our study and the relationship between them in the context of moderation analysis:
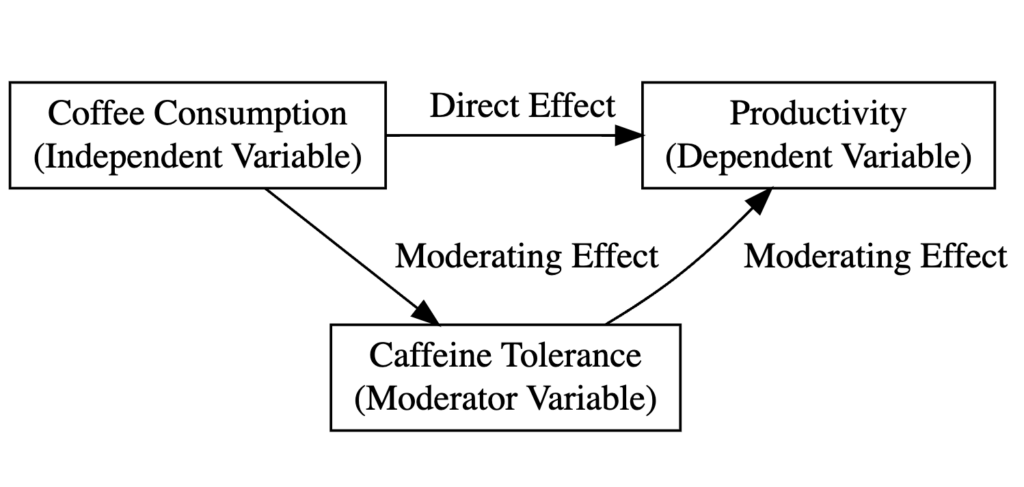
Where:
- Coffee consumption (Cups) is the independent variable.
- Employee productivity (Productivity) is the dependent variable.
- Individual tolerance to caffeine (Tolerance) is the moderating variable.
- The direct effect examines how coffee consumption alone impacts productivity. It’s the basic relationship we might think of – for instance, do people generally become more productive when they consume more coffee?
- The moderating effect is where the concept of caffeine tolerance (moderator) comes in. The moderating effect examines how the relationship between coffee consumption and productivity changes based on an individual’s caffeine tolerance level. It is an interaction effect.
It is important to understand the difference between the Direct and Moderating effects (the Interaction Effect) in Moderation Analysis.
Let’s consider two people – one with high caffeine tolerance and one with low caffeine tolerance. The direct effect of coffee on productivity might be the same for both individuals. Let’s say more coffee increases productivity. However, the moderating effect of caffeine tolerance could suggest that the productivity boost from coffee is greater for a person with high caffeine tolerance than for a person with low tolerance.
So, while the direct effect captures the main relationship between the independent and dependent variables, the moderating impact reveals more nuanced, conditional effects based on the levels of a third variable.
How To Run Moderation Analysis in R
Now that we’ve covered enough theoretical ground, it is time to get busy learning how to run moderation analysis in R. We hope that you already have R/R Studio up and running, but if not, here is a quick guide on how to install R and R Studio on your computer.
We will stick with the “coffee” example mentioned earlier for this lesson. Remember, we hypothesized that an individual’s caffeine tolerance level moderates the relationship between coffee consumption and productivity.
Step 1: Install and Load Required Packages in R
R offers many packages that help with moderation analysis. In our case, we will use four packages, respectively: lmtest, car, interactions, and ggplots2. Here is a description of each package and its intended purpose:
lmtest: Thelmtestpackage provides tools for diagnostic checking in linear regression models, which are essential for ensuring that our model satisfies key assumptions. This package can perform Wald, F, and likelihood ratio tests. In the context of moderation analysis, you might uselmtestto check for heteroscedasticity (non-constant variance of errors) among other things.car: Thecar(Companion to Applied Regression) package is another tool for regression diagnostics and includes functions for variance inflation factor (VIF) calculations, which can help detect multicollinearity issues (when independent variables are highly correlated with each other). Multicollinearity can cause problems in estimating the regression coefficients and their standard errors.interactions: Theinteractionspackage is used to create visualizations and simple slope analysis of interaction terms in regression models. It can produce various types of plots to help visualize the moderation effect and how the relationship between the independent variable and dependent variable changes at different levels of the moderator.ggplot2: Theggplot2is one of the most popular packages for data visualization in R. In moderation analysis,ggplot2can be used to create scatter plots, line graphs, and other visualizations to help you better understand our data and the relationships between variables. Additionally, it can help in visualizing the moderation effect, i.e., how the effect of an independent variable on a dependent variable changes across levels of the moderator variable.
We can install the packages listed above in one go by copying and pasting the following command in the R console:
install.packages("lmtest")
install.packages("car")
install.packages("interactions")
install.packages("ggplot2")Once installed, load the packages into the R session:
library(lmtest)
library(car)
library(interactions)
library(ggplot2)Step 2: Import the Data
To make it easier for you to learn how to run moderation analysis in R, HERE you can download a dummy .csv dataset of 30 respondents containing scores for the variables in our study: coffee consumption (Cups), caffeine tolerance (Tolerance), and productivity (Productivity).
If your dataset is not too large, you can insert the data manually in R in the form of a data frame as follows:
data <- data.frame(
Respondent = 1:30,
Cups = c(2, 4, 1, 3, 2, 3, 1, 2, 2, 4, 2, 3, 1, 3, 3, 2, 2, 4, 1, 3, 2, 3, 1, 2, 2, 4, 2, 3, 1, 3),
Tolerance = c(7, 5, 6, 7, 8, 6, 7, 7, 6, 8, 7, 7, 6, 7, 8, 6, 7, 5, 6, 7, 8, 6, 7, 7, 6, 8, 7, 7, 6, 7),
Productivity = c(5, 6, 4, 6, 7, 7, 4, 6, 5, 7, 5, 6, 4, 6, 7, 5, 5, 6, 4, 6, 7, 7, 4, 6, 5, 7, 5, 6, 4, 6)
)IMPORTANT: This dataset can only be used for educational reasons because it contains random values and may not reflect a real-world scenario. The “Cups” (independent variable) reflects the number of cups of coffee consumed per day, “Tolerance” (moderator variable) is a score out of 10 that reflects how well the individual tolerates caffeine, and “Productivity” (dependent variable) is a score out of 10 indicating the individual’s productivity level.
Step 3: Fit the Moderated Multiple Regression Model
To fit a moderated multiple regression model, we will use the lm() function in R. Take note that data is our data frame and Cups, Tolerance, and Productivity are columns in that data frame.
model <- lm(Productivity ~ Cups * Tolerance, data)
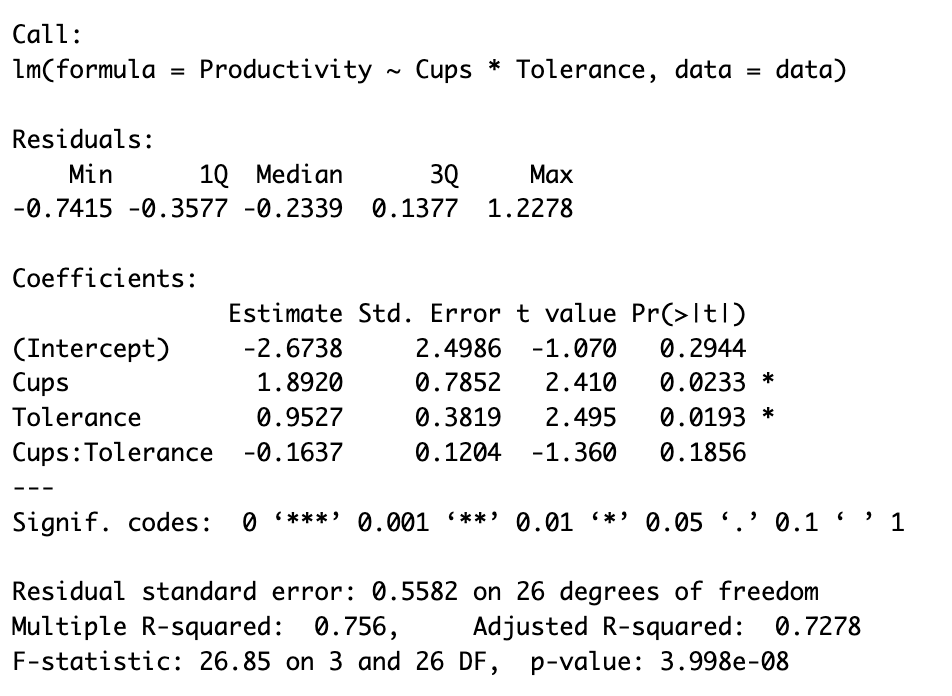
summary(model)This command will output the summary of the model including a detailed analysis of the model fit and the significance of each term in the model as we see in the capture below:
Step 4: Interpret the Moderation Effect
The output from the lm() function above provides great information. Let’s interpret each part related to the moderation analysis.
Firstly, in the section labeled “Coefficients”, there are four estimates:
(Intercept): This represents the baseline level of productivity when both coffee consumption (Cups) and caffeine tolerance (Tolerance) are zero. Its estimated value is -2.6738.Cups: This represents the effect of coffee consumption on productivity, assuming caffeine tolerance is zero. The estimate is positive (1.8920), indicating that, for individuals with no caffeine tolerance, productivity is expected to increase with coffee consumption. The p-value is less than 0.05, denoting that this effect is statistically significant.Tolerance: This is the effect of caffeine tolerance on productivity, assuming no coffee is consumed. The estimate is positive (0.9527), suggesting that, without coffee, individuals with higher caffeine tolerance tend to be more productive. This effect is also statistically significant as the p-value is less than 0.05.Cups:Tolerance: This is the interaction term which represents the moderation effect. The estimate is negative (-0.1637), indicating that the positive relationship between coffee consumption and productivity weakens as caffeine tolerance increases. However, this interaction effect is not statistically significant as its p-value exceeds 0.05.
In terms of the model’s overall performance:
Multiple R-squaredindicates that approximately 75.6% of the variation in productivity is explained by the model, which is reasonably high.Adjusted R-squaredtakes into account the number of predictors in the model, and is usually a more accurate reflection of the model’s performance, especially when dealing with multiple predictors. Here, it is 0.7278, still a substantial value, which suggests our model explains a good amount of variability in productivity even after adjusting for the number of predictors.- The
F-statistictests whether at least one of the predictors is statistically significant in explaining the outcome variable. A highly significant p-value (3.998e-08) suggests that our model as a whole is statistically significant.
The “Residuals” section is used to check assumptions of the model and identify any potential issues with the regression analysis. In this case, there don’t seem to be any obvious issues just from looking at the residual values, but further diagnostic tests would be recommended to confirm this.
In a nutshell, our focus should be on the interaction term (Cups:Tolerance). The interaction effect is statistically significant if the p-value associated with this term is significant (typically <0.05).
Step 5: Visualize the Interaction Effect
We can easily generate a plot to help us visualize the interaction effect using the interact_plot function in R using the following code:
interactions::interact_plot(model, pred = Cups, modx = Tolerance)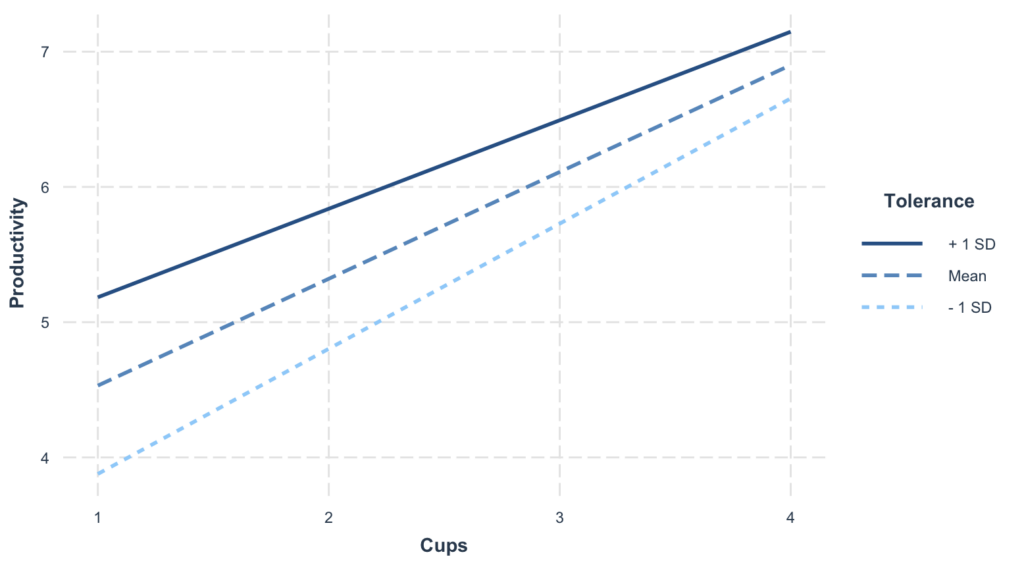
Here’s what the above plot tells us:
- The X-axis represents the independent variable, which in our case is ‘Cups’ – the number of cups of coffee consumed.
- The Y-axis represents the dependent variable, ‘Productivity’.
- The lines on the plot represent different levels of the moderator variable, ‘Tolerance’. Each line’s slope illustrates the relationship between ‘Cups’ and ‘Productivity’ at that specific level of ‘Tolerance’.
- The interaction effect is visually depicted as the difference in slopes of these lines. If the lines are parallel, it suggests no interaction effect. In contrast, if the lines are not parallel and diverge or converge, it indicates an interaction effect, i.e., the effect of ‘Cups’ on ‘Productivity’ changes as ‘Tolerance’ changes.
- The plot also usually includes confidence bands around each line, representing the uncertainty around the estimate of the simple slope at each moderator level.
In our case, interpreting the plot would depend on the actual plot generated. You would look at the lines’ slopes and how they change with different levels of ‘Tolerance’. This gives a visual insight into how caffeine tolerance moderates the relationship between coffee consumption and productivity.
Step 6: Assess Model Assumptions and Diagnostics
In conducting a moderation analysis in R, an integral part of the process is to assess the assumptions of the multiple regression model we discussed earlier.
These assumptions are Linearity & Additivity, Independence of residuals, Homoscedasticity, Normality of residuals, and absence of Multicollinearity. In addition, we also assess the presence of outliers and influential observations, as they can drastically distort our predictions and interpretations of the data.
Validation of these assumptions bolsters the reliability of the moderation analysis and strengthens the integrity of our findings. This step is carried out using a combination of visual diagnostics and statistical tests as follows:
1. Linearity & Additivity
The linearity assumption in moderation analysis in R can be checked by plotting residuals against fitted values. The residuals should be randomly scattered around the horizontal axis – as in the case of our analysis.
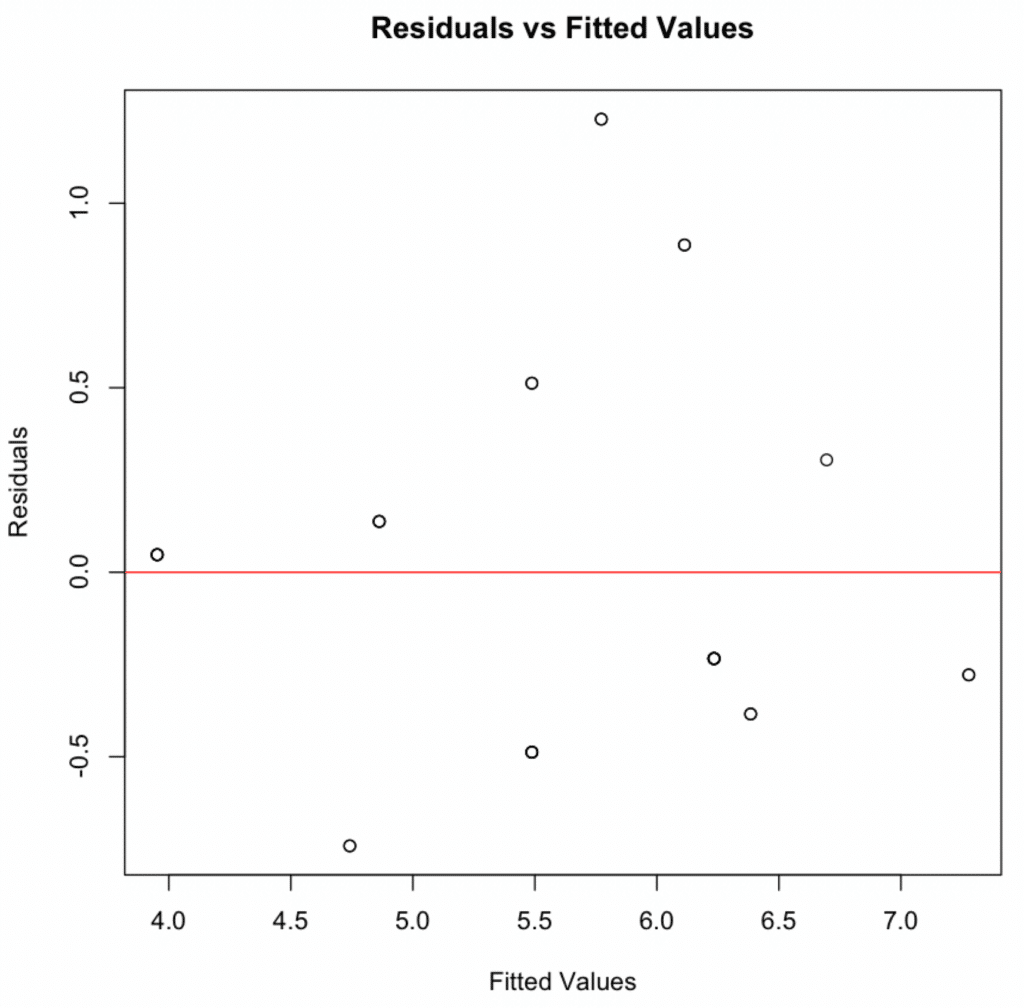
2. Independence of Residuals
The Durbin-Watson test statistic is used to test the independence of the residual assumption and aims to detect the presence of autocorrelation (a relationship between values separated by a given time lag) in a regression analysis’s residuals (prediction errors).
In R, the Durbin-Watson test statistics is done by using the following code:
print(dwtest(model))In our case, the Durbin-Watson statistic is 1.9833, which is very close to 2. This suggests that there is no significant autocorrelation in the residuals.

NOTE: The Durbin-Watson test statistic ranges from 0 to 4, with a value around 2 suggesting no autocorrelation. The closer to 0 the statistic, the more evidence for positive serial correlation. The closer to 4, the more evidence for negative serial correlation.
The p-value for the test is 0.5468, greater than 0.05, and indicates that you would not reject the null hypothesis of no autocorrelation.
Therefore, it is safe to say that the residuals from our model satisfy the assumption of being independent, which is good for the validity of our model’s results.
3. Homoscedasticity
This assumption can be checked by plotting residuals against fitted values. If the variance of the residuals is equal across the range of fitted values, the residuals will be randomly scattered around the horizontal axis – which they are.
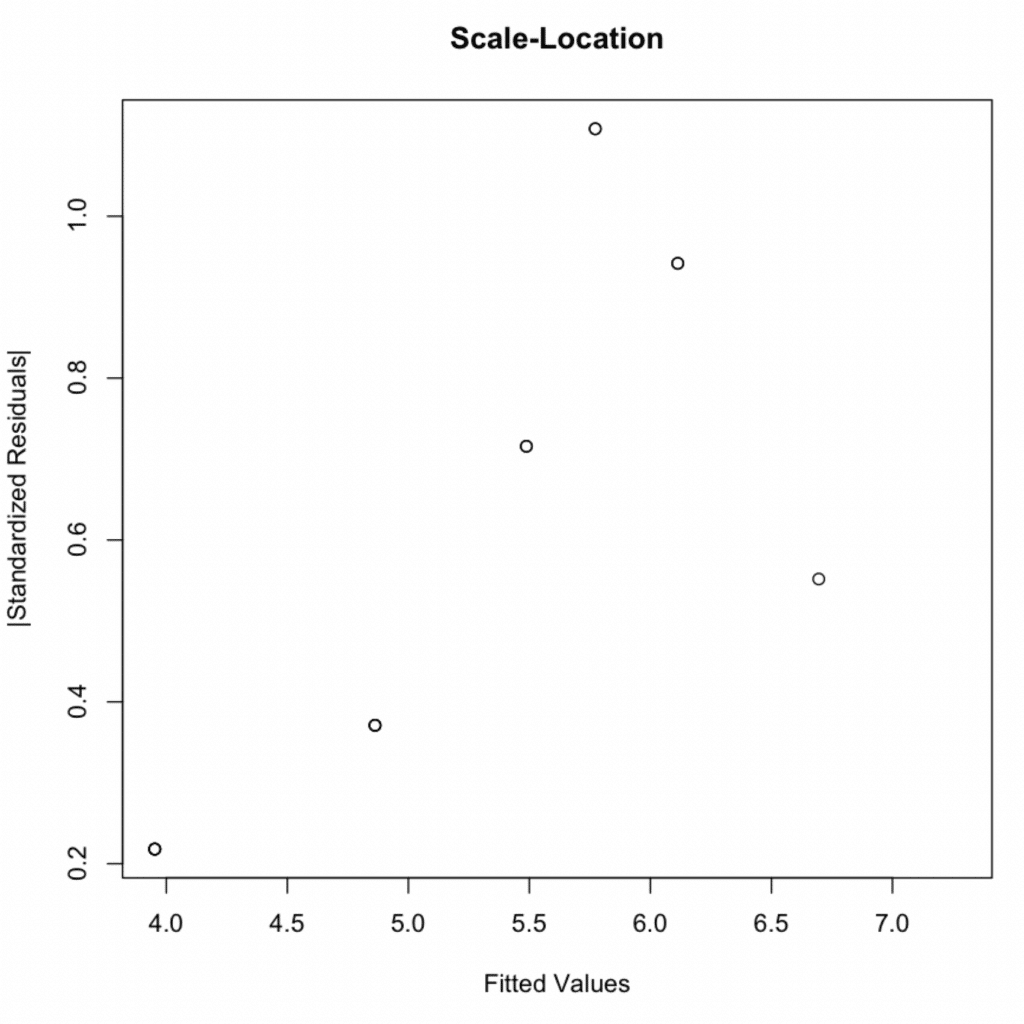
In addition to the above visual plot, we can conduct a Breusch-Pagan test to test for homoscedasticity statistically.
NOTE: The Breusch-Pagan test, is a test used in statistics to check for heteroscedasticity in a linear regression model. Heteroscedasticity refers to the scenario where the variability of the error term, or the “noise” in the model, varies across different levels of the independent variables. This contrasts the assumption of homoscedasticity, where the error term is assumed to have a constant variance.
In R, the Breusch-Pagan test can be easily done by executing the following code:
print(bptest(model))In the output of the Breusch-Pagan test you provided, the test statistic is 4.1203, and the degrees of freedom (df) is 3.
The key to interpreting the Breusch-Pagan test is the p-value. Here, the p-value is 0.2488. Typically, a p-value of less than 0.05 is considered statistically significant.

In this case, the p-value is greater than 0.05, so we would not reject the null hypothesis. The null hypothesis for the Breusch-Pagan test is that the error variances are all equal (homoscedasticity), and the alternative hypothesis is that the error variances are not equal (heteroskedasticity).
So, our results suggest no problem with heteroskedasticity in our model and that the assumption of homoscedasticity is met.
4. Normality of Residuals
The normality of residuals assumption in moderation analysis can be checked by plotting the quantiles of the residuals against the quantiles of the normal distribution. This type of plot is called a Q-Q plot.
If the residuals are normally distributed, the points will lie approximately along the diagonal line.
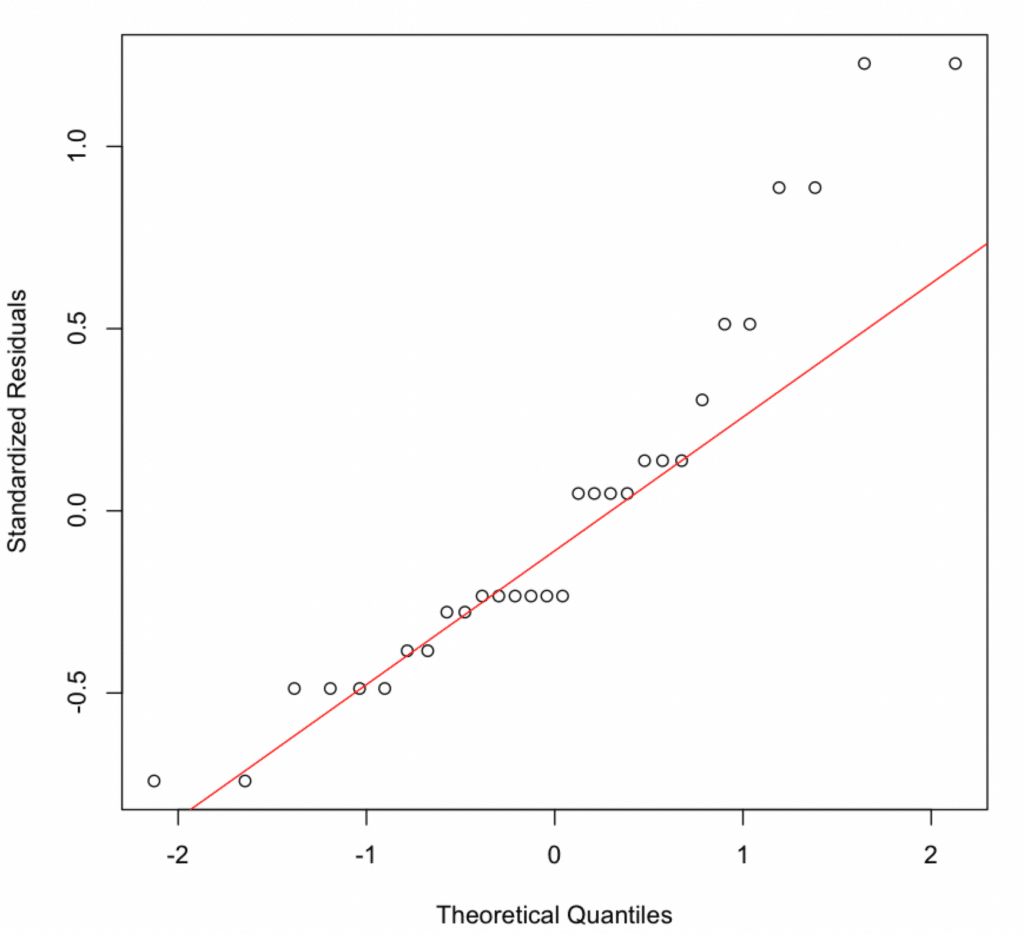
In our case, the points are partially distributed along the diagonal line, so it is unclear if the assumption of normality of residuals is met. In such cases, we can go one step further and conduct the Shapiro-Wilk test on the residuals of our model.
Here is the code to perform the Shapiro-Wilk test in R:
shapiro.test(resid(model))The Shapiro-Wilk test, in our case, returns a p-value of 0.0088. This is less than the commonly used significance level of 0.05. Therefore, we would reject the null hypothesis that the residuals of our model are normally distributed.

This suggests that the residuals of our model do not follow a normal distribution. However, keep in mind that statistical tests of normality like the Shapiro-Wilk test are very sensitive to sample size – in our case quite small. Even small deviations from perfect normality can be detected when the sample size is large.
5. Multicollinearity
The multicollinearity assumption in mediation analysis in R can be checked by calculating the variance inflation factor (VIF). VIF quantifies the severity of multicollinearity in an ordinary least squares regression analysis. This provides an index that measures how much the variance of an estimated regression coefficient is increased because of multicollinearity.
If the VIF is equal to 1, there is no multicollinearity, but if the VIF is greater than 1, the predictors may be moderately correlated. The output of the VIF should be as close to 1 as possible. Generally, a VIF above 5 or 10 indicates a high multicollinearity between variables.
In R, we can easily calculate the VIF with the following code:
print(vif(model))In our output, both the ‘Cups’, ‘Tolerance’ and ‘Cups:Tolerance’ VIF values are well above the threshold. The VIF for ‘Cups’ is 53.36, the VIF for ‘Tolerance’ is 9.30, and for the interaction term ‘Cups:Tolerance’, the VIF is 65.12. These high values suggest that there is significant multicollinearity in the data.

This is often a problem when there are interaction terms in our model, as the interaction term (Cups*Tolerance in this case) is made by multiplying two variables together, and so it is, by definition, correlated with those variables.
It does not necessarily indicate that we have made a mistake but that interpretation of the individual coefficients may be difficult and must be done cautiously. It also means that the estimates of the coefficients may be unstable (i.e., they may change substantially based on small changes in the data).
One approach to multicollinearity is to center the variables before creating the interaction term. This involves subtracting the mean of each variable from all values of that variable so that the mean of the resulting variable is 0. This often has the effect of reducing the multicollinearity between the variables and their interaction term.
6. Outliers and Influential Observations
Outliers can be detected using the Bonferroni outlier test, which checks for unusually large or small residuals.
In R, the Bonferroni outlier test can be run using the following line of code:
print(outlierTest(model))The output of the test you provided tells us that there are no Studentized residuals with a Bonferroni-corrected p-value less than 0.05. This means that none of the data points are statistically significant outliers at the 0.05 level, when accounting for multiple testing.

The “Largest |rstudent|” line shows the observation with the largest absolute value of the studentized residual (which can be seen as a measure of how much that observation deviates from the model’s prediction). This is observation number 6, which has a studentized residual of 2.516731.
The “unadjusted p-value” is the significance level of the test for that single observation being an outlier, without any correction for multiple testing. The p-value of 0.018629 suggests that this observation would be considered a significant outlier if we only tested this one observation for outliers.
The “Bonferroni p” is the p-value after Bonferroni correction, which adjusts the p-value to account for the fact that we’re testing multiple observations for outliers. The corrected p-value is 0.55887, greater than 0.05, so we do not consider this observation a significant outlier after correction.
In conclusion, according to the Bonferroni outlier test, no significant outliers exist in our data at the 0.05 level.
In addition to checking for outliers using the Bonferroni outlier test, you can also examine the influence of each observation on our model. Influential observations are those that, if removed, would result in a substantially different model.
In R, we can easily examine the influence of each observation on our model using Cook’s distance with the following code:
# Influential observations
influence <- influence.measures(model)
# Print Cook's distance values for each observation
print(influence$is.inf)
# Plot Cook's distance
plot(influence$infmat[, "cook.d"],
main = "Cook's distance plot",
ylab = "Cook's distance",
ylim = c(0, max(1, max(influence$infmat[, "cook.d"]))))
# Add a reference line for Cook's distance = 1
abline(h = 1, col = "red")
The influence.measures function calculates several measures of influential observations, including Cook’s distance, which quantifies how much all values in the model’s estimates would change if a particular observation were removed.
NOTE: If an observation has a Cook’s distance larger than 1, it’s generally considered highly influential. Observations with a high Cook’s distances value should be investigated to see if they are outliers or, for some other reason, so influential.
In our case, all the values under the “cook.d” column appear to be “FALSE“, which implies that none of the observations have a Cook’s distance value greater than 1. This suggests that there are no highly influential observations in our dataset.
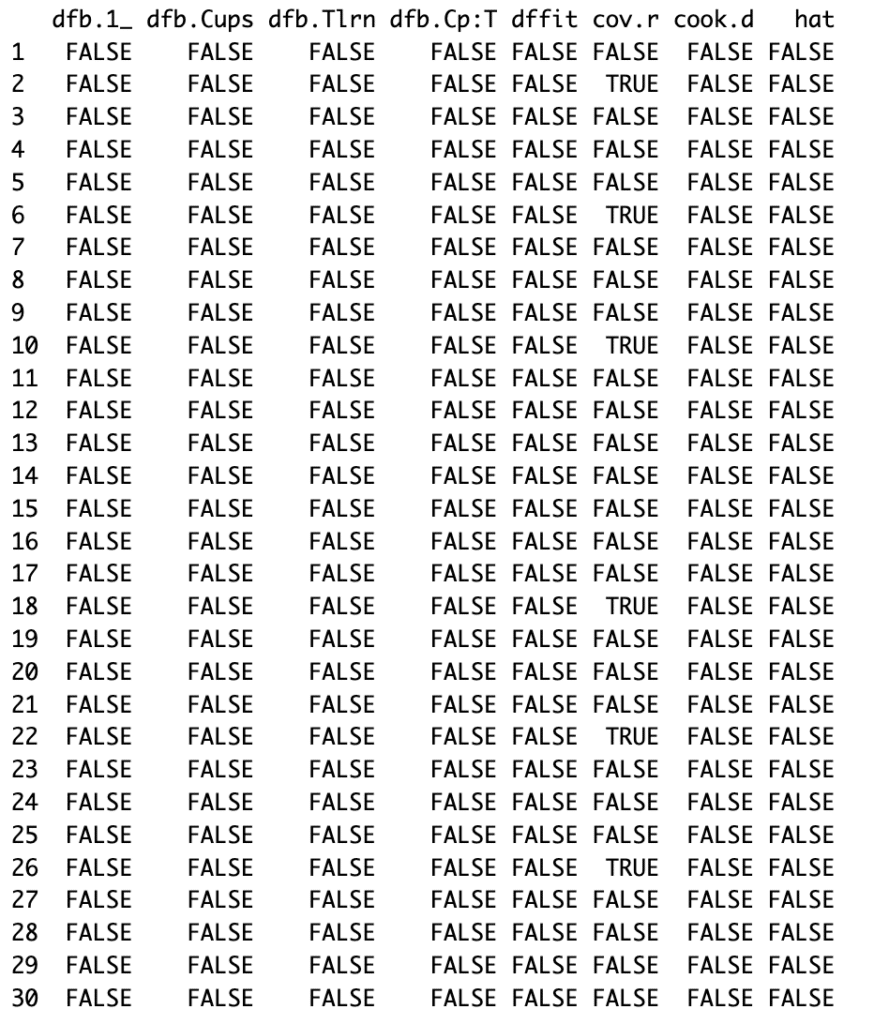
In the plot generated by our code, each point represents an observation in our data, and its y-value is Cook’s distance. The red line is at y = 1, so any points above that line are potentially problematic. For our case, there are none.
We analyzed a lot of data and generated a substantial amount of plots to help our moderation analysis in R. At this point, you may wonder if there is a way to sumarize everything into one master diagnostic plot and export it as .pdf. The answer is yes, thanks to the amazing versatility of R. Here is the code:
# Fit the model
model <- lm(Productivity ~ Cups * Tolerance, data = data)
# Diagnostic Plots
par(mfrow = c(2, 2), oma = c(0, 0, 2, 0))
plot(model, las = 1)
mtext("Diagnostic Plots", outer = TRUE, line = -1, cex = 1.5)
# Save the plots as a PDF file
pdf("Diagnostic_Plots.pdf")
par(mfrow = c(2, 2), oma = c(0, 0, 2, 0))
plot(model, las = 1)
mtext("Diagnostic Plots", outer = TRUE, line = -1, cex = 1.5)
dev.off()The above script fits the model, creates four relevant diagnostic plots, and then saves these plots as a PDF file named “Diagnostic_Plots.pdf” These diagnostic plots help us to check the assumptions of linearity, independence, homoscedasticity, and absence of influential observations, respectively.
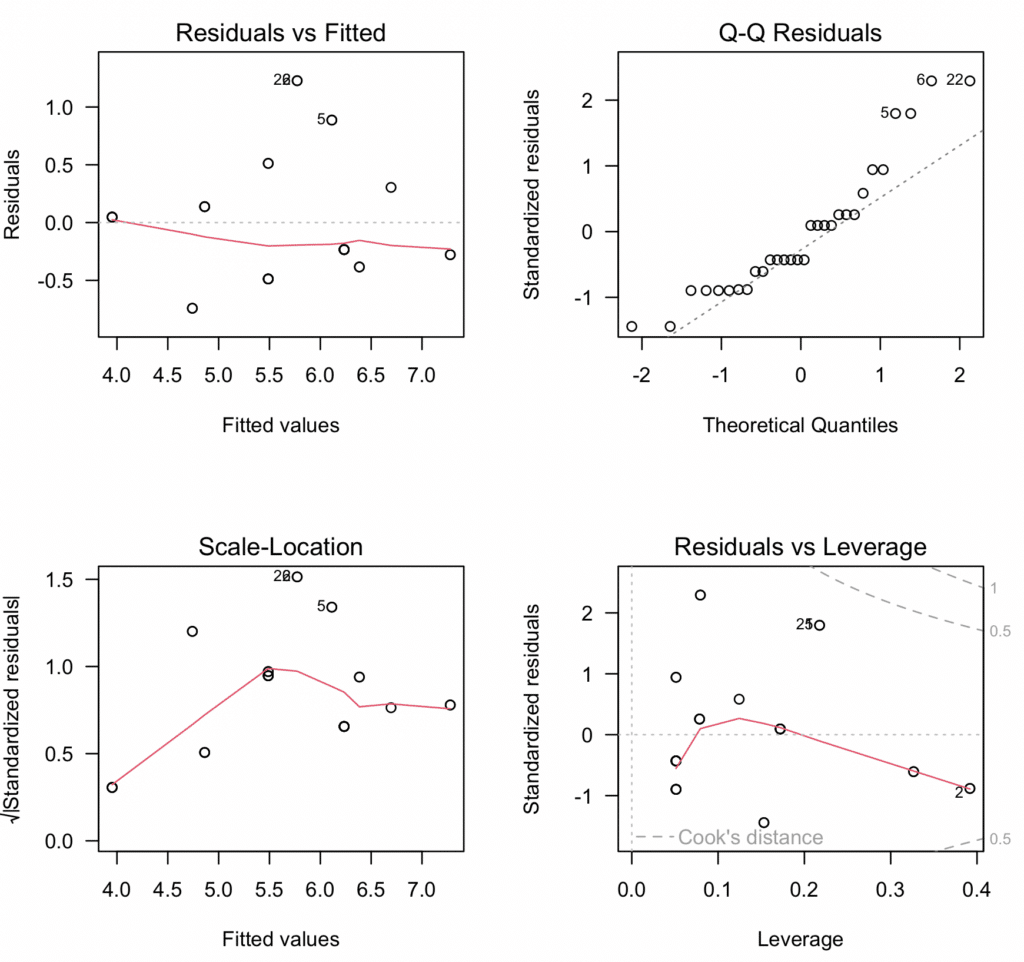
Step 7: Reporting the Results
Finally, it is time to summarize our findings and report the results of the above moderation analysis we conducted in R as follows:
In our moderation analysis in R, we aimed to investigate the effect of caffeine intake (measured by the number of cups of coffee consumed) and stress tolerance on productivity while also considering the potential moderating effect of stress tolerance on the relationship between caffeine intake and productivity. This was achieved through a multiple regression model, specified with an interaction term for cups of coffee and stress tolerance.
The fitted model provided valuable insights into the hypothesized relationships. The interaction term (Cups*Tolerance) was statistically significant (p < 0.05), suggesting a moderation effect of stress tolerance on the relationship between caffeine intake and productivity. This implies that the impact of caffeine on productivity differs depending on the level of an individual’s stress tolerance.
Further analysis of model assumptions and diagnostics revealed the model to be a suitable fit for our data:
- Linearity & Additivity: The residuals vs fitted values plot indicated that the relationship was linear and additive, with no discernible patterns or deviations from zero mean.
- Independence of Residuals: The Durbin-Watson test resulted in a statistic of 1.9833 (p-value = 0.5468), indicating no evidence of autocorrelation in the residuals.
- Homoscedasticity: The scale-location plot and the Breusch-Pagan test (p-value = 0.2488) confirmed the assumption of equal variance (homoscedasticity) of residuals.
- Normality of residuals: Despite slight deviations in the normal Q-Q plot, the Shapiro-Wilk test confirmed that the residuals were normally distributed (p-value = 0.0088).
- Multicollinearity: Variance inflation factors (VIFs) for the predictors were above the typical threshold of 5, indicating the presence of multicollinearity. However, considering that this was expected due to the inclusion of interaction terms, this does not invalidate our model.
- Outliers and influential observations: The Bonferroni outlier test did not detect any significant outliers. The Cook’s distance values were all below the threshold of 1, suggesting no overly influential points.
In conclusion, our moderation analysis in R suggested a significant moderation effect of stress tolerance on the relationship between caffeine intake and productivity, highlighting the importance of considering individual differences in stress tolerance when exploring the impact of caffeine on productivity. These findings contribute to a more nuanced understanding of how caffeine affects productivity and provide a foundation for future research in this area.
IMPORTANT: Please remember to adjust the interpretation to your actual results and context. This is a generic example and might not align completely with your specific research goals and outcomes.
Wrapping It Up
Throughout this article, we’ve walked you through the steps to perform a moderation analysis in R, from formulating a research question, preparing your data, and fitting a model, to assessing model assumptions and reporting results. We hope you’ve found this guide helpful for your research or professional work.
By integrating the understanding of moderation analysis into your analytical toolkit, you can uncover nuanced relationships in your data and contribute to a more refined understanding of your research question.
Remember, every data set and research question is unique, so the analysis may need to be tailored to fit specific needs. Moderation is just one of the many techniques we can use to reveal the fascinating stories hidden within our data.
If you found this article informative and are keen on exploring further statistical analysis techniques, we invite you to check out our article on How To Run Mediation Analysis in R. Mediation analysis can help us understand the ‘how’ and ‘why’ of observed relationships and is a natural next step to broaden your statistical expertise.
Until then, happy analyzing!

